Background: Mortality prediction models are increasingly being adopted in the clinical setting, both to retrospectively assess quality of care and to prospectively inform clinical practice. An open question is whether a particular hospital should employ a model trained using a diverse nationwide dataset or use a model developed primarily from local data. The Veterans Affairs (VA) healthcare system uses a national risk-adjusted mortality model for all inpatient hospitalizations for the purposes of retrospective performance assessment and improvement. This national model is developed using nationwide data and uses logistic regression with multiple covariates. The model is regularly re-fitted to prevent model degradation. In this study, we compare performance of this national mortality prediction model against two mortality prediction models developed using only single hospital data; we hypothesize that a mortality-prediction model using single hospital data may improve prediction of 30-day mortality compared to a model generated from a large national dataset.
Methods: This retrospective study included all patient admissions to the general medicine ward at VA Palo Alto Health Care System from October 1, 2018 to September 30, 2021. Patient data were extracted from the VA Corporate Data Warehouse (CDW). Covariates were the same as the national model and included age, marital status, admission diagnosis, admission source, Elixhauser comorbidity diagnoses, and comprehensive metabolic laboratory values. Both linear regression and a machine-learning-based approach (gradient boosting) were used. The H20 AutoML package was used to generate both local models. Testing was performed in a temporal fashion with three months of observation data tested on the subsequent three months of data. The national model was prebuilt with predicted 30-day mortality stratified into five categories: < 2.5%, 2.5%-5%, 5%-10%, 10%-30%, and >30%. For each model type, we evaluated performance for varying training set sizes using the receiver operator characteristic area under curve (ROC-AUC) as the benchmark performance metric.
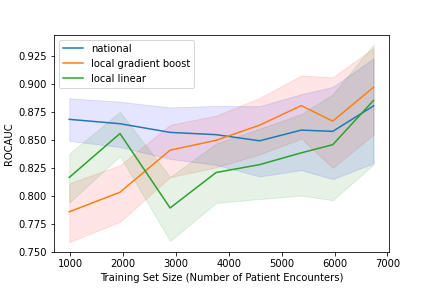
Results: A total of 8,261 patient admissions were included representing 5,732 unique patients. The local linear regression model was inferior to the national model until training sets included 18 months of data and 3,500 admissions at which point the two models performed comparably. With 24 months of data representing 5,000 admissions, the machine learning model outperformed the national model though with overlapping confidence intervals. In the gradient boosting model, patients admitted to non-medicine services were low predicted risk for mortality. Age was proportional to predicted risk of mortality. All forms of metastatic cancer were high projected risk of mortality. Low laboratory albumin and elevated blood urea nitrogen levels were highly predictive of 30-day mortality.
Conclusions: For single-site clinical or operational use, machine learning models using only local data may provide more accurate predictions of 30-day mortality than larger multi-site models. Other benefits of using local data may include ease of operational integration for clinical decision support, speed of re-fitting, and model customization. Future research should prospectively evaluate the impact of such predictions toward improvement of clinical outcomes.