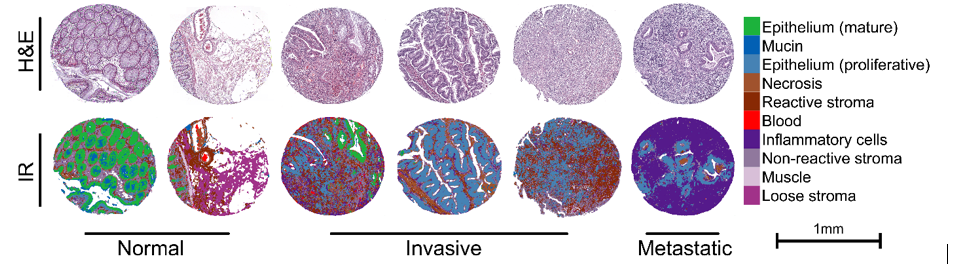
Background: The tumor microenvironment and its interaction with the tumor are indicative of disease progression but has not been clinically applied due to the lack of objective assessment criteria. Proximity associated effects of the tumor to its microenvironment which can explain the long-term behavior of tumor remains to be developed. In this work, we utilized the biochemical sensitivity of infrared spectroscopic imaging to segment colon tumor biopsies into major tumor microenvironment components.
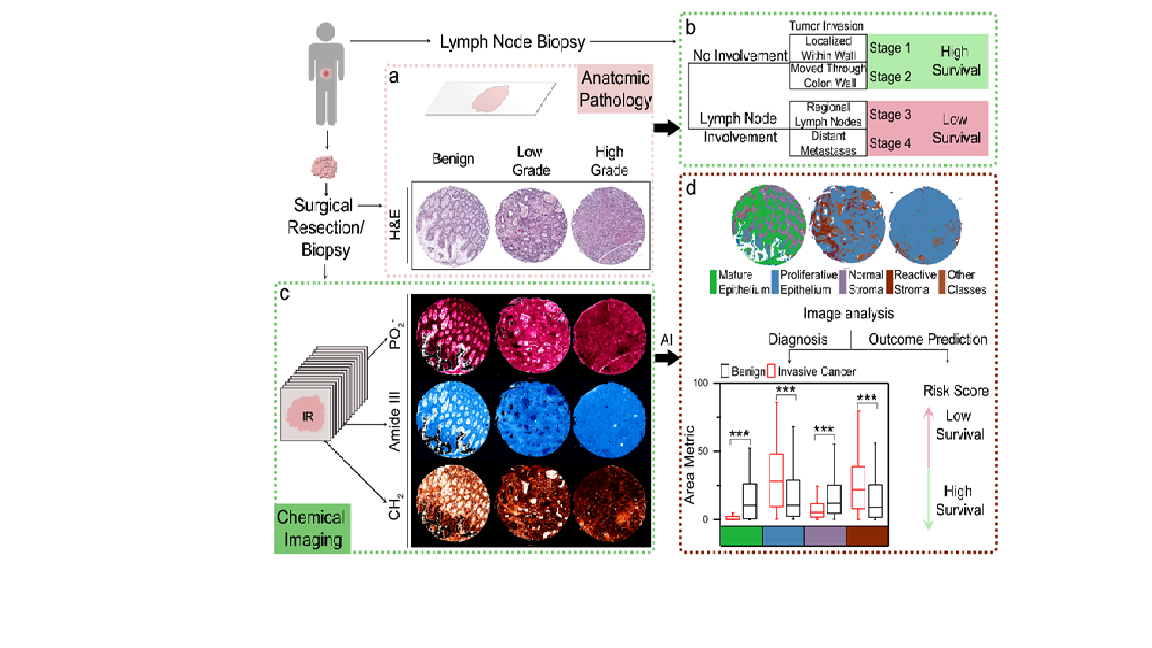
Methods: Our patient cohort comprised of 320 anonymized patients who underwent surgical resection for colorectal carcinoma. Cores of 1 mm diameter were sampled for each patient from representative invasive areas and were used to construct eight tissue microarrays(TMA). Data was annotated by labeling histologic classes on H&E images. These annotations were then manually copied on the corresponding infrared spectroscopic images as well. The spectral signatures associated with the desmoplastic reaction, normal stroma and lymphocytes, and seven other tissue components were obtained by matching regions on the H&E image to the infrared spectroscopic images. Post ANOVA and mRMR analysis, we retained 50 spectral metrics that were used for developing the histology classifier model.
Results: Cell populations and micro-environmental features showed differences in their infrared absorbance spectra owing to the inherent biochemical differences between them. Mucin and mature epithelium clustered close together, while blood had the most distinctive features, owing to the absence of nucleic acids. In invasive tumors, the glandular structure formed by malignant epithelium is distorted or lost, and the stroma around the tumor is modified due to the desmoplastic reaction (reactive stroma). All of these histologically important features were classified correctly. We were also able to identify the presence of malignant cells in samples from lymph node metastases, indicating satisfactory performance. Spectral profiles collected from IR imaging enabled sensitive and specific identification of tumor and tumor microenvironment components which can be mapped onto the tissue using artificial intelligence(AI) for robust image quantification. Three risk-associated features were defined to assess the tumor-stroma interaction. Three different risk variables were analyzed which were distance metric (the distance of the malignant epithelium to the stroma), area metric ( radius of the stromal component) and interaction metric ( interaction of the previous mentioned two features for each pixel). The univariate and multivariate cox-regression analysis showed a direct association of poor overall survival and cancer aggressiveness with reactive stroma(desmoplasia).
Conclusions: A bio-molecular snapshot of the tumor in its native microenvironment captured in conjugation with the spatial distribution could be descriptive of the patient outcomes. Our work on quantitative chemical imaging[CMI] can provide a robust approach to capture and model the biochemically specific spatial interactions to predict long-term tumor behavior. IR imaging-based quantitative image analysis can also provide key insights into several other cancer types, especially in cases where outcome associated molecular markers are lacking. We anticipate applying quantitative chemical imaging approaches in clinics, allowing automated diagnosis and outcome predictions thus leading to more standardization as well as rapid results.